Application Note
CRISPR Electroporation of T Cells Improves Treatment in Patients with Refractory Cancer
Background
Cancer immunotherapy is a fast-moving field that blends cutting-edge technologies to overcome past obstacles in the clinic. A key challenge has been refractory cancers which are advanced in progression or have not responded to prior therapies. Clinicians have intervened with adoptive T cell therapy, engineering the patient’s own T cells to enhance the body’s natural anti-tumor immune response to combat malignancies.
Unfortunately, even sophisticated treatment modalities are vulnerable to familiar complications such as T cell exhaustion and autoimmunity, or, as seen with early chimeric antigen receptor T cell (CAR T) studies, an increased risk of Cytokine Release Syndrome (CRS). To improve the safety and efficacy of adoptive T cell therapies, methods have been introduced to use T Cell receptors (TCRs) instead of CAR T to mitigate the risk of CRS.
MaxCyte® offers an immune cell, gene editing solution to help improve treatment efficacy and lead to better clinical outcomes. CRISPR ribonucleoproteins (RNPs) can knockout functional genes, such as TRAC and TRBC, limiting TCR mispairing and improving the expression of cancer-specific, transgenic TCRs. Similarly, CRISPR-mediated deletion of the immune check point inhibitor PD-1 has been shown to limit T cell exhaustion.
MaxCyte Gene Editing in an Efficient T Cell Manufacturing Workflow
Aim
The groundbreaking study presented here was designed to develop and assess the safety and feasibility of an improved immunotherapy using NYCE (NY-ESO-1-transduced CRISPR 3X edited cells). Featured experiments test the ability of CRISPR-Cas9 editing, enabled by clinical scale electroporation with the MaxCyte ExPERTTM platform, to modify T cells for sustained ability to attack and kill tumor cells while maintaining patient safety.
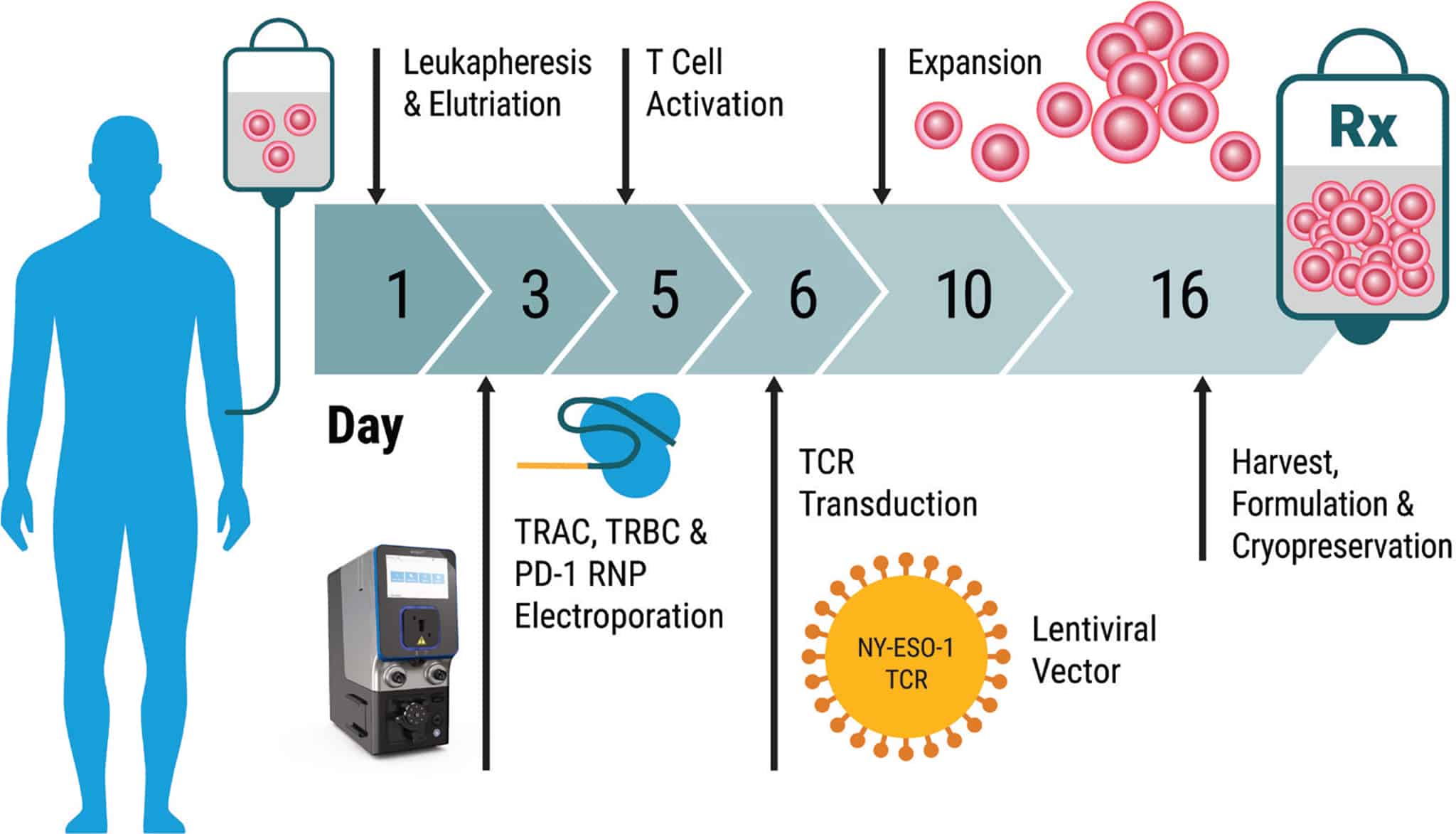
Method Overview: Electroporation of Patient Cells at Clinical Scale with the MaxCyte GTx®
Outlined below is a summary of the 16-day protocol, from patient cell harvest through manufacturing of the final cell therapy product. This illustrates the speed with which cancer immunotherapies can be made and the seamless fit of MaxCyte enabling technology into an efficient T cell manufacturing process.
Day 1 • Harvest – Autologous patient cells were obtained by apheresis, enriched for lymphocytes, and seeded in culture.
Day 3 • CRISPR Gene Editing – Patient T cells were processed through MaxCyte electroporation to deliver CRISPR RNPs with an equimolar mixture of Cas9 and sgRNAs targeting TRAC, TRBC and PD-1. Cells were incubated at 30° C for two days to enhance gene editing efficiency.
Day 5 • T Cell Activation – Cells were shifted to 37° C and activated using CD3/CD28 beads.
Day 6 • Lentiviral Transduction – Cells were transduced with NY-ESO-1 TCR and maintained in static culture.
Day 10 • Expansion – Cells were expanded in a WAVE bioreactorTM. A minimum of 1x1010 cells per patient were collected upon harvest.
Day 16 • Harvest – Beads were removed and NYCE cells were formulated and cryopreserved until patient infusion.
Full methods for cell handling and patient protocols are detailed in Science 28 Feb 2020: Vol. 367, Issue 6481.
Results
Safer Therapy Through Gene Editing
Cells collected from four cancer patients presenting myeloma or sarcoma were engineered with a combination of MaxCyte electroporation followed by lentiviral transduction. Briefly, cells were expanded and edited to knockout TRAC, TRBC and PDC1 with gene modification efficiencies ranging from 15-45% in the final infusion products (Figure 1). While off-target effects of CRISPR are a key concern that clinicians continue to monitor, in this study over 90% of editing occurred on-target and patients receiving therapeutic infusions did not develop humoral responses to Cas9 protein (Table 1).

Figure 1. High Fidelity Gene Editing at 3 loci. The frequency of gene-disrupted total cells in NYCE infusion products was measured using chip-based digital PCR. All data are representative of at least two independent experiments. Error bars represent mean + SEM.
Table 1. Measurement of On-Target Editing Efficiency for Each Gene by Final Product


Figure 2. Potency of Final Engineered NYCE Cells Before Infusion. NYCE cells were co-cultured with HLA-A2+ tumor cells engineered to express NYESO-1 and luciferase. Patient T cells transduced with the NY-ESO-1 TCR without CRISPR-Cas9 editing (labeled NY-ESO-1 TCR) and un-transduced T cells with CRISPR-Cas9 editing of TRAC, TRBC, and PDCD1 (labeled CRISPR) were included as controls (n = 4 patient T cell infusion products). Asterisks indicate statistical significance determined by paired Student’s t tests between groups (*P <0.05). Error bars represent SEM.
Enhanced Potency and Persistence
Initial estimates of NYCE potency were obtained by co-culture with tumor cells expressing target antigen. NYCE cells had potent, antigen-specific cytotoxicity over a wide range of effector-to-target ratios when compared to controls that were either transduced with only the TCR or electroporated with CRISPR RNP alone (Figure 2).
When a chip based digital PCR approach was used to measure edited cell number before and after patient treatment, 5–10% engraftment frequency of TRAC and PD-1 knockout cells was achieved (Figure 3).
Improved Phenotype and Stability of NYCE T Cells in Patients
Three patients were treated with lymphodepleting chemotherapy followed by a single infusion of NYCE cells. For four months following infusion, NYCE cells were evaluated in patient blood samples, at intervals, by single cell RNA sequencing to determine transcriptomic phenotype. Uniform manifold approximation and projection (UMAP) plots of gene expression confirmed stable frequencies of NYCE gene-edited cells from 10 days to four months post infusion (Figure 4).

Figure 3. In Vivo Persistence of NYCE Cells. Chip-based digital PCR was used to measure frequencies of CRISPR-Cas9–edited T cells (TRAC, TRBC, and PDCD1 knockout) before and after adoptive cell transfer. Error bars represent SD.
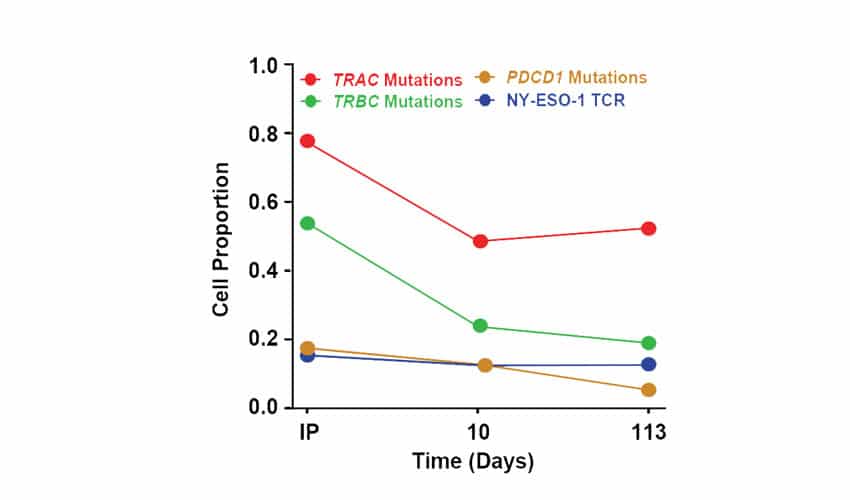
Figure 4. Transcriptomic Phenotype and Evolution of NYCE T Cells in Patients. Single cell RNA sequencing was used to characterize the transcriptomic phenotype of the NYCE cells and their evolution over time in one patient. Engineered cells were infused into this patient and blood was collected at d10 and 4 months after infusion. T cells were sorted and processed using droplet 5’scRNAseq. Proportions of pre-infusion (IP, day 0) and post infusion (days 10 and 113) wild-type T cells with TRAC, TRBC, or PDCD1 mutations or expressing the NY-ESO-1 TCR transgene.
Verified Therapeutic Efficacy and Patient Response After Infusion of NYCE Cells
To determine if engineered cells modified with MaxCyte electroporation retained their antitumor activity after infusion, peripheral blood mononuclear cell (PBMC) samples were collected from patients at 3-9 months after infusion and expanded in vitro in the presence of NYESO-1 peptide and the cytokine interleukin-2 (IL-2) which promotes general proliferation and function of T cells (Figure 5A).
The ability of expanded immune effector cells to recognize antigen and elicit cytotoxicity against target cells was tested in a chromium release assay, and antigen specific cytotoxicity was observed in all three patients. Patient UPN39 (far right, Figure 5A) had the highest antitumor activity.
Finally, therapeutic efficacy was visible in patient UPN39 whose CT scan shows 50% regression of a large abdominal mass after infusion of NYCE cells (Figure 5B).


Figure 5. Cytolytic Activity and Patient Outcomes After Infusion of NYCE Cells. A) PBMC samples were collected from patients at 3, 6 or 9 months after infusion. Cells were expanded in vitro in the presence of NYESO-1 peptide and IL-2. The ability to recognize antigen and elicit cytotoxicity against target cells was tested in a 4h 51Cr release assay incorporating Nalm-6 NY-ESO-1+, parental Nalm-6 (NY-ESO-1−), and A375 melanoma cells (NY-ESO-1+). B) CT scans show 50% tumor regression of a large abdominal mass in patient UPN39 after infusion.
Conclusion and Future Applications
In summary, this is the first ever, human clinical trial to demonstrate multiplex CRISPR gene editing of T cells from patients with advanced, refractory cancer, resulting in a durable, safe and effective TCR immunotherapy. Therapeutic T cells, modified with the MaxCyte ExPERT platform, engrafted and survived for months in the human body, a significant improvement over many previous approaches where these cells lost their function within days.
In addition to having sustained ability to attack and kill tumors, the infusions were well tolerated, with no cytokine release syndrome or detected reaction to Cas9 protein. The result is a treatment that is not only more effective but safer for the patient.
The study presented here is an important milestone towards the ultimate goal of helping the body’s own immune system recognize and attack cancer, but the promise of the approach extends far beyond this. In fact, there are thousands of cell and gene therapy clinical trials ongoing worldwide to investigate modification of immune and stem cells for disease treatment.2 With the demonstrated safety and efficiency of multiplex gene editing and MaxCyte enabling technology, we now have not only a strong foundation for genome engineering but the potential for future improvement of innovative medicines.
References
- CRISPR-engineered T cells in patients with refractory cancer. (2020) Science367(6481): eaba7365.
- Gene & Cell Therapy FAQs: ASGCT. www.asgct.org/education/more-resources/gene-and-cell-therapy-faqs